Related: Bioinformatics MOC, 10 X Genomics Dataset
Go to the desired paper with available data - check on the right-hand side for a title called “Data availability.” This should take you to section in the paper with a link to an online website where the data is hosted.
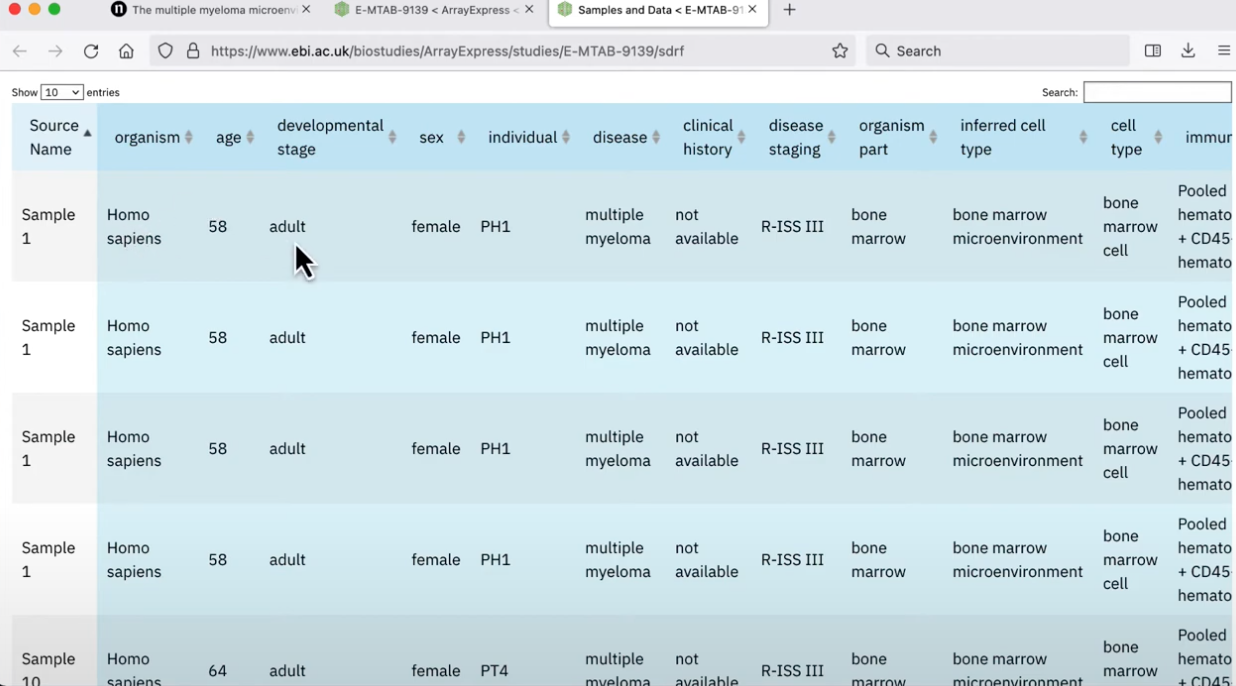
checking for meta-data
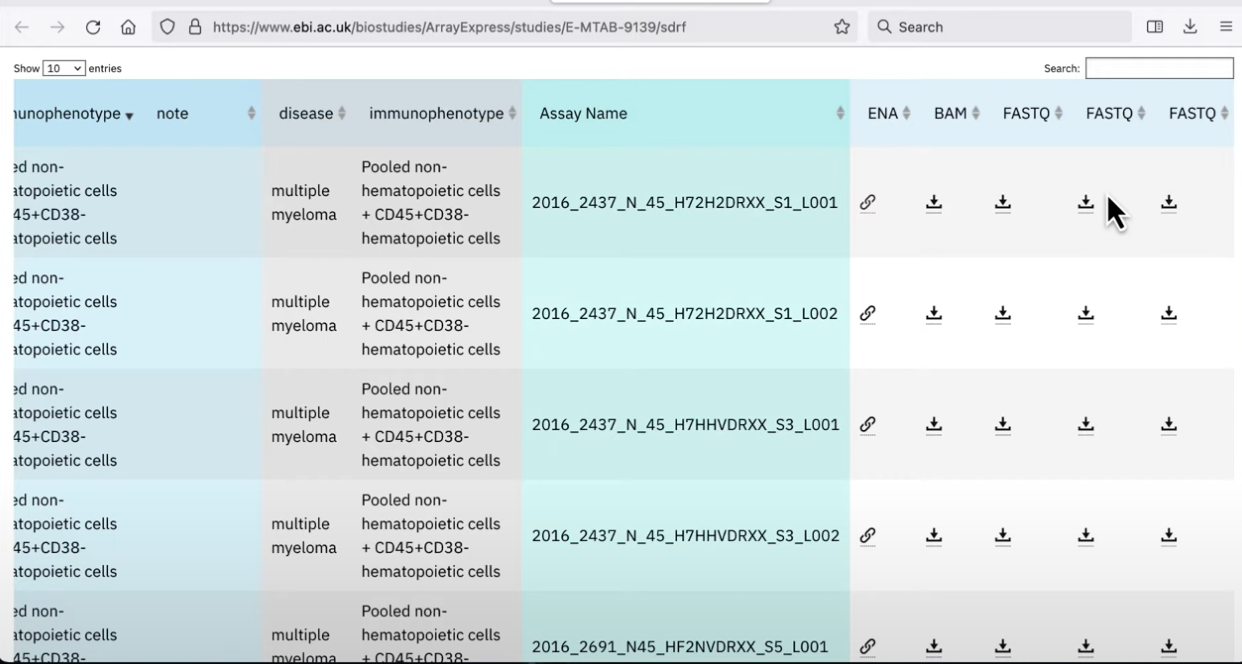
There should be a place to download ALL the data, but then also a table which describes where the data is coming from. For example a table that lists all of the samples, whether they are normal controls or have a disease, meta-data about the dataset, and the separate FASTQ files
 |  |
|---|---|
| downloading data to a remote server via a terminal |
- open terminal
- create a folder per 1 subject
- go back to the meta-data on the user-interface and click “copy link” on the dataset you want to download (have to do this one at a time if it’s multiple)
- run wget (Linux) and then Cmd-V to paste link that you just copies
- now it’s being remotely downloaded and will show you how long it will take
- run ls in the downloaded directory to check if it’s there, you can also check using Finder
if you don’t have wget, you can get it via homebrew or use curl on Mac
via homebrew install
brew install wgetvia curl
curl [url] -o [name-your-file]pre-processing step via cellranger
run the cellranger library from 10x genomics prepocesses scrnaseq data count on the videos